Scientific and Central Projects
Information and contact details
CP1: Translational platform for PDAC models and drug response validation
In order to study the mechanistic, functional and therapeutic consequences of subtype-specific and subtype-overlapping genome dynamic alterations in PDAC, the consortium relies on molecularly characterized patient-derived PDAC models suitable for comprehensive applications ranging from detailed mechanistic studies to pre-clinical drug testing. To this end, CP1 has established a translational PDAC model platform comprising Patient-Derived-Xenograft (PDX) models, PDX-derived primary PDAC cells (CDX) and patient-derived-organoid (PDOs) models for joint utilization within and beyond the CRU5002. PDAC models are generated from PDAC patients enrolled in the Molecular Pancreas Program (MolPAC) of the UMG and reflect all disease stages. In cooperation with CP2, we conduct thorough molecular characterization of the primary PDAC specimen and the translational models at the genomic and expression (transcriptome and immunohistochemistry) level. Further, CP1 functions as the interface of the CRU5002 and the Molecular Tumor Board (MTB) of the UMG and benefits from the MTB pipeline as a temporally distinct second source of PDAC samples and models. Finally, CP1 offers an in vitro PDAC model platform for basal functional characterization of PDO and CDX models and standardized and quality-controlled assessment of drug (combination) responses. Together, CP1 envisions to substantially support the CRU5002 in dissecting the biological significance and the therapeutic consequences of genome dynamics in PDAC and to speed up the translational potential of CRU5002 findings.
PIs:
Elisabeth Heßmann, Klinik für Gastroenterologie, gastrointestinale Onkologie und Endokrinologie
Matthias Dobbelstein, Institut für Molekulare Onkologie
Silke Kaulfuß & Prof. Dr. Bernd Wollnik, Institut für Humangenetik
Philipp Ströbel, Institut für Pathologie
Team:
Jennifer Appelhans (Ströbel Group)
Christin Kellner (Hessmann Group)
Jessica Spitalieri (Hessmann Group)
Sercan Mercan (Ellenrieder Group)
Joana Oschwald (Hessmann Group)
Sebastian Schimkowiak (Dobbelstein Group)
Jovan Todorovic (Ströbel Group, GEROK position)
Lennart Versemann (Hessmann Group)
Chan Rong Lai (Schneider Group)
Claudia Hartmann (Schneider Group)
If you are interested in our translational PDAC models, please contact elisabeth.hessmann(at)med.uni-goettingen.de.
CP2: Biomedical Informatics Support Platform (BISP)
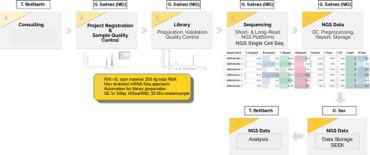
The Biomedical Informatics Support Platform (BISP) of CP2 supports the CRU5002 translational research efforts through three key components:
1. Development of infrastructure to streamline the management of both clinical and research data (U. Sax).
2. Implementation of cutting-edge sequencing methods and advanced analytical techniques (G. Salinas).
3. Provision of a wide range of bioinformatics pipelines and expert guidance to enable effective data analysis and integration (T. Beissbarth).
These objectives are achieved through the implementation of the following:
1. A SEEK-based system that efficiently manages and provides accessible data to all CRU5002 members, alongside a local cBioPortal platform for seamless integration of experimental and clinical data.
2. The establishment of innovative single-cell omics methods, specifically designed and optimized for PDAC samples, which have overcome previous technical limitations and opened up new avenues for PDAC research.
3. The integration of state-of-the-art bioinformatics techniques, focusing on PDAC subtyping, genome dynamics, and gene regulatory networks, which have significantly enhanced CRU5002’s capabilities and provided exciting opportunities for future advancements.
Our research focuses on comprehensively characterizing genomic similarities and differences between primary PDAC, PDO, PDX, and CDX models. This focus will continue on the integration of functional data from translational PDAC models, while continually maintaining and refining centralized platforms and NGS methods. We are also committed to setting new standards and methods for data generation and integrative data analysis, including consulting and training for these platforms.
Additionally, we are developing new sequencing strategies in single-cell omics and expanding data analysis pipelines for gene regulatory networks and genome dynamics. As part of our goal to translate CRU5002 findings into clinical practice, another major focus is the integration of experimental and clinical data.
Therefore, CP2 not only facilitates collaborations within the SPs and with CP1 but is also essential for the successful implementation of CRU5002’s ultimate aim: stratification-based therapy driven by genome dynamics.
PIs:
Tim Beißbarth, Institut für Medizinische Bioinformatik, UMG
Gabriela Salinas, NGS - Integrative Genomics Core Unit (NIG), UMG
Ulrich Sax, Institut für Medizinische Informatik, UMG
Team:
AG Sax:
Nils Beyer
Sophia Rheinländer
AG Beißbarth:
Martin Haubrock
Martin Stoves
AG Salinas:
Jaqueline Fink
Fabian Ludewig
Susanne Luthin
Further information on the Translationale Verbundforschung, the Department of Medical Bioinformatics and the NGS - Integrative Genomics Core Unit (NIG) is available on their respective websites.
Internal Links:
Fairdom SEEK
The central data management and storage platform of the KFO5002. All data files are uploaded here to make them findable for the members of the KFO, and accessible with a detailed roles and rights management.
URL: https://seek.kfo5002.umg.eu
Accessibility: UMG WissLan (VPN instructions: contact CP2)
Login: LDAP login (UMG Account without domain)
SharePoint
Collaboration platform where Microsoft Office files are edited simultaneously.
URL: Contact Elisabeth Hessmann and CP2 for access
Accessibility: Internet
Login: LDAP login (UMG account with domain, e.g. UMG\mustermann)
cBioPortal
Data query and analysis platform with harmonized data for every CRU patient integrated from diverse sources.
URL: https://cbioportal.kfo5002.umg.eu
Accessibility: UMG WissLan (VPN instructions: contact CP2)
Login: UMG Account without domain
SP1: Characterizing genome dynamics in ARID1A-deficient PDAC subtypes
38% of PDAC are characterized by alterations in genes encoding chromatin regulatory proteins, particularly members of the SWItch/Sucrose Non-Fermentable (SWI/SNF) family. Given the widespread loss-of-function mutations of different subunits of the SWI/SNF complex in a plethora of malignancies, members of this chromatin remodeling complex are referred to as pivotal tumor suppressors. AT-rich Interactive Domain-containing protein 1A (ARID1A) is the most frequently altered subunit of the SWI/SNF complex in PDAC and has been associated with an aggressive PDAC phenotype and a bad disease outcome. Importantly, in other tumor entities, ARID1A-deficiency is connected to increased sensitivity towards PI3K/AKT-, ATR-, PARP- HDAC- or EZH2-inhibition, thus suggesting assessment of the ARID1A status as a promising therapy-predictive strategy in cancer treatment. However, whether ARID1A deficiency also predicts for certain therapeutic vulnerabilities in PDAC remains mainly elusive. SP1 aids at filling this gap and utilizes the structural and material-based resources as well as the diverse scientific expertise of the CRU5002 to explore the mechanistic background and the functional implications of genome dynamic alterations that particularly characterize ARID1A-deficient PDAC subtypes. Moreover, we investigate the therapeutic consequences of ARID1A-status specific genome dynamic alterations and hence sought to elucidate whether and how the ARID1A status can be considered for stratification-based therapy in PDAC treatment.
PI: Elisabeth Hessmann, Department for Gastroenterology, Gastroinestinal Oncology, and Endocrinology, University Medical Center Göttingen
Team:
Christin Kellner
Denise Schlösser
Ankit Goswami
Jiska Ziegler
Further information on the AG Heßmann is available on their website.
SP2: Exploiting oncogenic transcription factor complexes in misp53 PDAC subtypes
The high aggressiveness as well as the chemoresistance of PDAC are significantly determined by the diverse genomic alterations occurring throughout disease development and progression. More than 75% PDAC tumors exhibit genetic alterations in TP53 tumor suppressor functions. In particularly, missense TP53 mutations are more prevalent among the PDAC patients with high-grade and metastatic cancer. However, the extent to which these missense TP53 mutants determine unfavorable prognosis and therapeutic outcome in PDAC remains unclear. SP2 aids at gaining mechanistic insights into distinct missense TP53 mutants and their consequences on genome dynamic alterations driving PDAC aggressiveness and therapy resistance.
PIs:
Ramona Schulz-Heddergott, Institut für Molekulare Onkologie, UMG
Shiv K. Singh, Klinik für Gastroentreologie, gastrointestinale Onkologie und Endokrinologie, UMG
Team:
Working Group Schulz-Heddergott:
Jessica Grabowsksi
Juline Nehring
Kim-Lucia Schneider
Working Group Singh:
Rebecca Samuel
Xueyen Wu
Nehir Sahin
Jian Shi
Frederike Penz
Further information on the Schulz-Heddergott Group and the AG Singh is available on their respective websites.
SP4: GSK3βhigh subtype-specific DNA repair mechanisms and resistance in PDAC
GSK3β is a multifunctional serine/threonine kinase, which due to its broad substrate specificity, is involved in numerous cellular processes, e.g. glucose metabolism, proliferation and stem cell identity. Changes in GSK3β expression, localization and kinase activity have been reported in various human diseases, e.g. type-2 diabetes mellitus, Alzheimer’s disease and cancer. In cancer, GSK3β can exert both tumor suppressor and promoter functions depending on the cellular and molecular context. In addition to the well-established tumor suppressor functions of GSK3β, increasing evidence also supports oncogenic activities of the kinase in various epithelial and non-epithelial malignancies, e.g. mixed lineage leukemia, glioblastoma and oral cancer, in which activation of the kinase promotes cancer growth and metastasis as well as chemoresistance. We and others have univocally shown the powerful oncogenic properties of GSK3β-signaling activation in pancreatic cancer (PDAC), where it favors acquisition of a poorly differentiated and highly aggressive phenotype. Within SP4, we want to further understand its functions, especially focusing on the involvement of GSK3β in DNA-repair mechanisms. Hereby is of special interest how different epigenetic alterations can influence this pathway and serve as a predictor of the GSK3β inhibition in PDAC therapy. The platform of the KFO 5002 provides with its multiple models the necessary basis to evaluate the potential of GSK3β inhibition in the stratification of patients.
PI: Volker Ellenrieder, Klinik für Gastroentreologie, gastrointestinale Onkologie und Endokrinologie, UMG
Team:
Aiko Bockelmann (AG Ellenrieder)
Madhubanui Dey (AG Ellenrieder)
Margo Dudek (AG Ellenrieder)
Pascal Hoffmeister (AG Ellenrieder)
Íñigo Vicente Hernandez (AG Ellenrieder)
Further information on the AG Ellenrieder is available on their website.
SP5: Metabolic impact on chromatin topology in subpopulation-derived PDAC organoids
A hallmark of both tumorigenesis and cancer progression is the metabolic reprogramming of cancer cells, and an interest in targeting metabolic cancer traits has resurged. Specifically, as regards pancreatic cancer, the TCGA (The Cancer Genome Atlas) classification of PDACs was recently found to align well with distinct metabolic states. Classical PDAC tumors mostly rely on lipogenic metabolism, while the more aggressive squamous/ mesenchymal PDACs predominantly rely on glycolysis, with the two subtypes also displaying striking differences in glucose and glutamine utilization, as well as in mitochondrial function. These differences suggest differential sensitivity of PDACs to inhibitors targeting glycolysis, glutamine metabolism, lipid synthesis or redox balance. This coincidence between PDAC subtypes and metabolic states becomes even more relevant once one considers that specific metabolic pathways directly affect global chromatin methylation and acetylation levels and may, thus, contribute to the dramatic dysregulation of epigenetic profiles in tumours. It is exactly this link that our project will investigate: by combining studies of spatial genome organization, cis-element mapping, and single-cell transcriptomes in patient-derived PDAC organoids we aim at uncovering regulatory circuits that can be targeted for therapeutic intervention.
PIs:
Lena Conradi, Klinik für Allgemein, Viszeral- und Kinderchirurgie, UMG
Argyris Papantonis, Institut für Pathologie, UMG
Team:
AG Conradi:
Tiago De Oliveira
Birgit Jünemann
Henrik Spahn
Tobias Tacke
Sara Younes
Fabio Gätje
Alexandra Schmitt
Kelvin Lai
Santhiya Rajan
AG Papantonis:
Markos Tsitsianopoulos
Further information on the AG Papantonis and the AG Conradi is available on their respective websites.
SP6: Causes and consequences of SMAD4- and TGFβ-regulated PDAC genome instability
Whole chromosome instability (W-CIN) is a hallmark of cancer and is defined as an increased rate of chromosome missegregation during mitosis leading to evolving aneuploidy. By perpetually altering the genetic composition of cancer cells W-CIN can fuel clonal evolution of tumors, tumor progression and the development of therapy resistance. The sub-project 6 focuses on the role of TGFβ signaling for the induction of W-CIN in pancreatic cancer. In particular, we hypothesize that PDAC subgroups characterized by high TGFβ signaling and by loss of SMAD4 are prone for the induction of chromosomal stability, which supports tumor progression, tumor aggressiveness and therapy resistance. The sub-project 6 will systematically investigate the presence and the mechanisms of W-CIN in PDAC, which will lead to a first definition of PDAC sub-types characterized by mitotic errors and W-CIN. Furthermore, we aim to elucidate the nature of mitotic errors present in chromosomally unstable PDACs, both in cell lines and in PDX-derived cells. By interfering with TGFβ signaling we will develop experimental routes to suppress CIN in PDAC cell models to directly address the question for the role of CIN in acquiring aggressive tumor phenotypes and in mediating resistance towards clinically relevant therapy regimens.
PI: Holger Bastians, Institut für Molekulare Onkologie, Sektion Zelluläre Onkologie, UMG
Team:
Dr. Simran Kaur (AG Bastians)
Gargee Joshi (AG Bastians)
Further information on the Bastians Group is available on their website.
SP7: Synergistic PDAC cell ablation by inhibiting CDKs and oncogenic signaling
Besides classical chemotherapy, the use of cyclin dependent kinase (CDK) inhibitors for treating pancreatic cancer is subject to intense investigation. Our project explores the efficacy of drug combinations that comprise such inhibitors. We are testing the hypothesis that inhibitors of cyclin dependent kinase 4 (CDK4) can eliminate PDAC cells and tumors in synergy with additional inhibitors that target oncogenic signalling pathways or chromatin modifiers. This hypothesis addresses a drug synergy, similar to our research during the first funding period (Ewers et al., 2021). It is based on our previous findings that the depletion or inactivation of signalling components perpetuates the cell cycle arrest imposed by inhibitors of CDK4. Mechanistically, the cell cycle is arrested by CDK4 inhibition. Under most circumstances, this arrest is only transient. Simultaneously interfering with oncogenic signaling, however, permanently precludes further proliferation. We are now planning to assess the genetic tumor cell specificities that determine the degree of this drug synergy, comparing PDAC-derived cell lines, patient-derived cells and organoids of the CRU5002 cohort. In this context, we are assaying the transcriptome and genome-wide chromatin modifications. Taken together, the project aims at improving PDAC therapy with CDK4 inhibitors, through the identification of synergistic drug combinations.
PI: Matthias Dobbelstein, Institut für Molekulare Onkologie
Team:
Maj-Britt Paulsohn
Further information on the Institute of Molecular Oncology is available on their website.
SP8: Exploiting the Transcription Cycle to Tailor Rational-Based Combination Therapies for PDAC
The high failure rate of anti-cancer drugs underscores the need to develop context-specific therapies. In pancreatic ductal adenocarcinoma (PDAC), cancer dependency scores identified the catalytically active subunit of protein phosphatase 2A (PP2A) as a specific target. Increased PP2A expression correlates with a poorer prognosis and is involved in metabolic, inflammatory, and proliferative signaling pathways. Using a clinically tested PP2A inhibitor, we were able to show that inhibition of PP2A disrupts the regulation of RNA polymerase II (RNA Pol II), leading to CDK9-mediated transcription elongation and subsequent cell death in mesenchymal PDAC cells. To further investigate the molecular mechanisms of PP2A inhibitor sensitivity, we are pursuing a multifactorial approach combining pharmacological inhibition of PP2A with genetic gain- and loss-of-function models. In addition, we analyze the influence of glycolysis and develop PP2Ai-based combination therapies.
PI: Günter Schneider, Department for General-, Visceral-, and Pediatric Surgery, UMG
Team:
Rong Chan Lai
Nicole Rjasanow
Atharva Naik
Ningjun Duan
Further information on the Schneider Group is available on their website.
You may also be interested in
