Our concept
Genome Dynamics in Pancreatic Cancer: Deciphering Mechanisms and Vulnerabilties for Precise Therapies
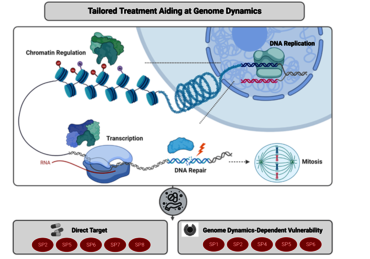
Pancreatic ductal adenocarcinoma (PDAC) remains a major challenge in cancer medicine with a desperate need to develop better treatment strategies. In fact, despite significant research efforts, PDAC still displays the highest mortality rate among all solid tumors in mankind. Major causes for its devastating disease outcome are the exceptionally aggressive tumor biology and the remarkable resistance to conventional anti-tumor treatments. Both characteristics are mechanistically linked to a very high degree of tumor heterogeneity, which is reflected by recent identification of various molecular and phenotypic PDAC subtypes. Importantly, compelling evidence from translational studies further revealed that multiple subtype-specific features are mediated by disturbed genome dynamics — e.g., defects in genomic stability or altered chromatin regulation and transcription. Therefore, understanding how genome dynamics drive subtype-specificity in PDAC progression and resistance will provide unique and novel avenues to refine PDAC treatment strategies towards stratification-based precision medicine.

The major goal of this Clinical Research Unit (CRU 5002) is to define the mechanisms, functional relevance and therapeutic potential of altered genome dynamics in selected PDAC subtypes. This initiative builds upon the different and highly complementary scientific and methodological expertise in PDAC patient care, basic research as well as bio- and medical informatics of the 16 contributing principal investigators of the University Medical Center Göttingen (UMG). The seven highly interlinked scientific projects (SP) of this CRU are directly supported by two essential central projects (CP) providing preclinical tumor models as well as platforms for sequencing technologies and biomedical informatics. In addition, this CRU 5002 tremendously benefits from the expanding scientific and clinical infrastructure at the Göttingen Campus and has unlimited access to a large amount of patient tumor material and corresponding clinical annotations received from the interdisciplinary Molecular Pancreatic Cancer Program (MolPAC) at the UMG.
We anticipate that our multidisciplinary and collaborative research consortium will not only improve our understanding of subtype specificity in PDAC biology and therapy resistance, but will also create solid foundations for stratification-based tailored treatment strategies in PDAC.
You may also be interested in